Abstract
Predicting disease progression remains a particularly challenging endeavor in chronic degenerative disorders and cancer, thus limiting early detection, risk stratification, and preventive interventions. Here, profiling the spectrum of chronic myeloproliferative neoplasms (MPNs), as a model, we identify the blood platelet transcriptome as a proxy strategy for highly sensitive progression biomarkers that also enables prediction of advanced disease via machine learning algorithms. Using RNA sequencing (RNA-seq), we derive disease-relevant gene expression in purified platelets from 120 peripheral blood samples constituting two time-separated cohorts of patients diagnosed with one of three MPN subtypes at sample acquisition - essential thrombocythemia, ET (n=24), polycythemia vera, PV (n=33), and primary or post ET/PV secondary myelofibrosis, MF (n=42), and healthy donors (n=21).
The MPN platelet transcriptome reveals an incremental molecular reprogramming that is independent of patient driver mutation status or therapy and discriminates each clinical phenotype. Differential markers in each of ET, PV and MF also highlight candidate genes as potential mediators of the pro-thrombotic and pro-fibrotic phenotypes in MPNs. In ET and PV, a strong thromboinflammatory profile is revealed by the upregulation of several interferon inducible transmembrane genes (IFITM2, IFITM3, IFITM10, IFIT3, IFI6, IFI27L1, IFI27L2), interleukin receptor accessory kinases/proteins (IRAK1, IL15, IL1RAP, IL17RC) and several solute carrier family genes (SLC16A1, SLC25A1, SLC26A8, SLC2A9) as glucose and other metabolic transport proteins, and coagulation factor V (F5). In MF, fibrosis-specific markers were identified by an additional focused comparison of MF patients versus ET and PV, showing increased expression of several pro-fibrotic growth factors (FGFR1, FGFR3, FGFRL1), matrix metalloproteinases (MMP8, MMP14), vascular endothelial growth factor A (VEGFA), insulin growth factor binding protein (IGFBP7), and cell cycle regulators (CCND1, CCNA2, CCNB2, CCNF).
Also, focusing on the JAK-inhibitor ruxolitinib/RUX-specific signatures, we not only confirm previous observations on its anti-inflammatory and immunosuppressive effects (e.g. downregulation in our RUX-treated cohort of IL1RAP, CXCR5, CPNE3, ILF3) but also identify new gene clusters responsive to RUX - e.g. inhibition of type I interferon (e.g. IFIT1, IFIT2, IFI6), chromatin regulation (HIST2H3A/C, HIST1H2BK, H2AFY, SMARCA4, SMARCC2), epigenetic methylation in mitochondrial genes (ATP6, ATP8, ND1-6 and NDUFA5), and other proliferation, and proteostasis-associated markers as putative targets for MPN-directed therapy.
Mechanistic insights from our data highlight impaired protein homeostasis as a prominent driver of MPN evolution, with a persistent integrated stress response. Preliminary ex vivo data on MPN patient bone-marrow-derived CD34+ cells and cultured megakaryocytes validate a proteostasis-focused subset of our peripheral platelet RNA-seq signatures.
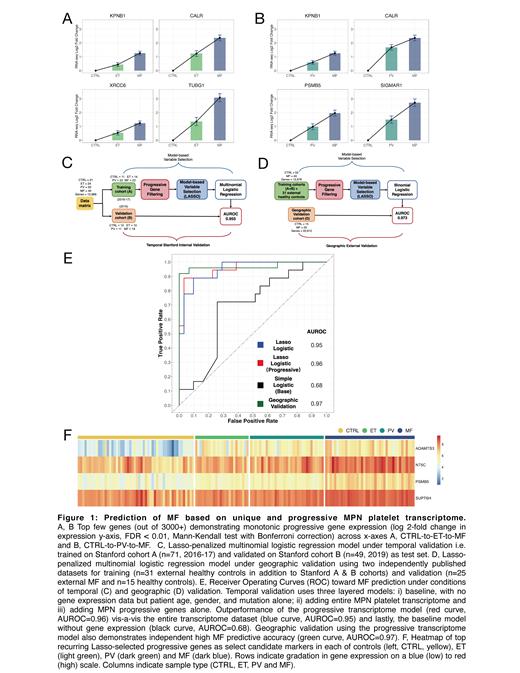
Further leveraging this substantive dataset, and in particular a progressive expression gradient across MPN, we develop a machine learning model (Lasso-penalized regression) predictive of the advanced subtype MF at high accuracy and under two conditions of validation: i) temporal Stanford internal (AUC-ROC of 0.96) and ii) geographic external cohorts (AUC-ROC of 0.97, using independently published data of an additional n=25 MF and n=46 healthy donors). Lasso-derived signatures offer a robust core set of < 5 MPN progression markers.
Together, our platelet transcriptome snapshot of chronic MPNs demonstrates a methodological avenue for disease risk stratification and progression beyond genetic data alone, with potential utility in a wide range of age-related disorders.
Part of the work contributing to this abstract has been posted as a preprint at this link: https://www.biorxiv.org/content/10.1101/2021.03.12.435190v2
Gotlib: Blueprint Medicines: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Incyte: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Deciphera: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Novartis: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Abbvie: Membership on an entity's Board of Directors or advisory committees, Research Funding; BMS: Research Funding; Kartos: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; PharmaEssentia: Honoraria, Membership on an entity's Board of Directors or advisory committees; Cogent Biosciences: Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Chair for the Eligibility and Central Response Review Committee, Research Funding; Allakos: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal